

Author: Ariel J. Ben-Sasson
Corresponding author: David Baker, Emmanuel Derivery
Correspondence: University of Washington, Cambridge MRC Molecular Biology Laboratory
Research highlights:
1. Developed a new method for computationally controlling the assembly of binary protein two-dimensional materials.
2. Realize the construction of an ordered two-dimensional array on a disordered substrate.
Two-dimensional materials are promising. Even biologists have begun to become interested in two-dimensional materials.
Ordered two-dimensional protein materials are composed of flexible links forming a single lattice, and these links are often composed of only one protein component. Naturally, scientists feel that a material composed of two protein components has more advantages.
Compared with materials composed of two or more components, genetically programmable materials that spontaneously co-assemble into an ordered structure after mixing two or more components are easier to control. It is easier for scientists to control assembly dynamics, perform strict characterization and manipulation of parts, and achieve more complex functions, so that the system can be used in various applications.
The two-dimensional ordered array of binary components has been confirmed, but due to the flexibility of the components, the structure of the design material cannot be completely determined in advance, and the construction unit has dihedral symmetry, and the array has the same upper and lower surfaces . The de novo design between rigid domains stabilized through a wide range of non-covalent interactions will provide more control over the atomic structure and provide a reliable starting point for further structural and functional adjustments.
In view of this, David Baker of the University of Washington and Emmanuel Derivery of the MRC Molecular Biology Laboratory in Cambridge and others reported a calculation method that can generate a co-assembled binary layer by designing a rigid interface between pairs of dihedral protein building units. So as to realize the two-dimensional superlattice of binary protein.
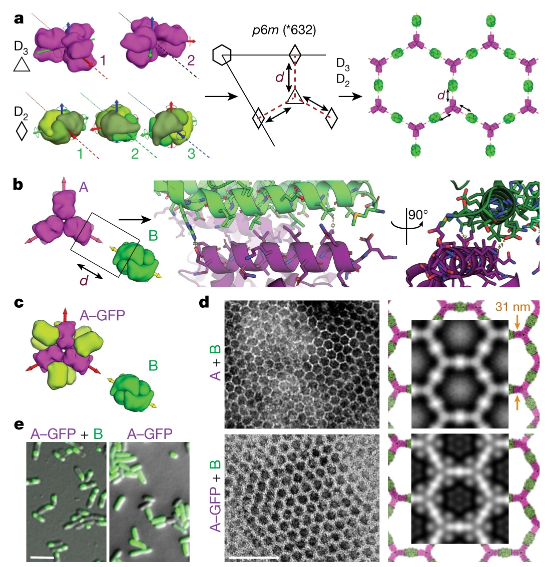
Figure 1. Design strategy
The array elements designed by the researchers will dissolve at millimolar concentrations. Interestingly, when they are combined at nanomolar concentrations, they will quickly assemble into almost crystalline micron-scale arrays without the need for two-dimensional material support. It is consistent with in vitro and intracellular computational design models.
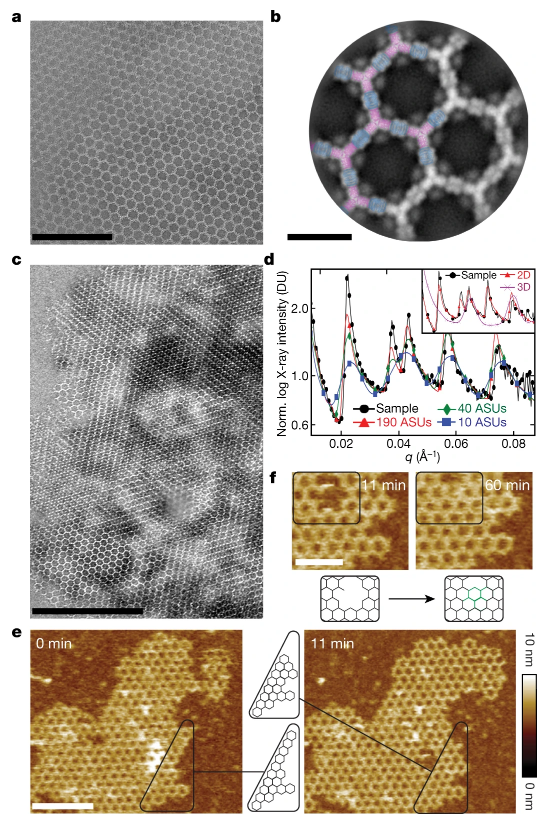
Figure 2. Assembly structure
Since the material is designed from the ground up, it is easy to functionalize the building units and reconstruct their symmetry to form a ligand array with distinguishable surfaces. Using atomic force microscopes and quantitative microscopes, the researchers found that the arrays assembled on the membrane have a stoichiometry and structure similar to those formed in vitro. This indicates that it is possible to achieve orderly assembly on a disordered substrate like the cell membrane.
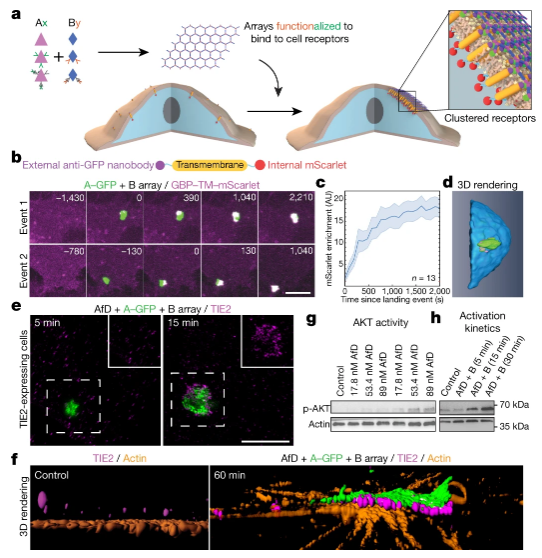
Figure 3. Kinetics
Previous studies have found that cell surface receptor binding elements (such as antibodies and nanocages) are quickly endocytosed. In contrast, the large array assembled on the cell surface this time can inhibit endocytosis in a tunable manner, which has the potential therapeutic significance of expanding receptor participation and evading immunity.

Figure 4. Mass assembled array
In short, this research has laid the foundation for synthetic cell biology and opened a new door for the development of two-dimensional materials!
references:
Ariel J. Ben-Sasson et al. Design of biologically active binary protein 2D materials. Nature 2020.
https://www.nature.com/articles/s41586-020-03120-8









